Data Tidiness
Overview
Teaching: 20 min
Exercises: 10 minQuestions
How to collect and structure the data about your sequencing data
Objectives
Think about and understand the types of metadata a sequencing experiment will generate.
Understand the importance of metadata and potential metadata standards
Explore common formatting challenges in spreadsheet data
Introduction
When we think about the data for a sequencing project, we often start by thinking about the sequencing data that we get back from the sequencing center, but just as important, if not more so, is the data you’ve generated about the sequences before it ever goes to the sequencing center. This is the data about the data, often called the metadata. Without the information about what you sequenced, the sequence data itself is useless.
Discussion
With the person next to you, discuss:
What kinds of data and information have you generated before you send your DNA/RNA off for sequencing?
Solution
Types of files and information you have generated:
- spreadsheet or tabular data with the data from your experiment and whatever you were measuring for your study
- lab notebook notes about how you conducted those experiments
- spreadsheet or tabular data about the samples you sent off for sequencing. Sequencing centers often have a particular format they need with the name of the sample, DNA concentration and other information.
- lab notebook notes about how you prepared the DNA/RNA for sequencing and what type of sequencing you’re doing, e.g. paired end Illumina HiSeq. There likely will be other ideas here too. Was this more information and data than you were expecting?
All of the data and information just discussed can be considered metadata, data about the data. We want to follow a few guidelines for metadata.
Notes
Notes about your experiment, including how you prepared your samples from sequencing, should be in your lab notebook, whether that’s a physical lab notebook or electronic lab notebook. For guidelines on good wet lab notebooks, see the Howard Hughes Medical Institute “Making the Right Moves: A Practical Guide to Scientifıc Management for Postdocs and New Faculty” section on Data Management and Laboratory Notebooks.
Including dates on your lab notebook pages, the samples themselves and in any records about those samples helps you associate everything with each other later. Using dates also helps create unique identifiers, because even if you process the same sample twice, you don’t usually do it on the same day, or if you do, you’re aware of it and give them names like A and B.
Unique identifiers
Unique identifiers are a unique name for a sample or set of sequencing data. They are names for that data that only exist for that data. Having these unique names makes them much easier to track later.
Once you start running computational analyses on your sequencing data, it’s just as important to keep a good computational lab notebook. Aside from usually being electronic, these are very similar to wet lab notebooks in that it’s best to be as detailed as possible. See Schnell’s “Ten Simple Rules for a Computational Biologist’s Laboratory Notebook” for more details on how to keep a useful computational notebook .
Data about the experiment
Data about the experiment is usually collected in spreadsheets, like Excel.
What type of data to collect depends on your experiment and there are often guidelines from metadata standards.
Metadata standards
Many fields have particular ways that they structure their metadata so it’s consistent and can be used across the field.
The Digital Curation Center maintains a list of metadata standards. Ones particularly relevant for genomics data are from the Genomics Standards Consortium.
If there aren’t metadata standards already, you can think about what the minimum amount of information someone would need to know about your data to be able to work with it, without talking to you.
Structuring data in spreadsheets
Independent of the type of data you’re collecting, there are standard ways to enter that data into the spreadsheet, to make it easier to analyze later. We often enter data that makes it easy for us as humans to read and work with it, because we’re human! Computers need data structured in a way that they can use it, so to use this data in a computational workflow, we need to think like computers when we use spreadsheets.
The cardinal rules of using spreadsheet programs for data:
- Put each observation or sample in its own row.
- Put all your variables in columns - the thing that vary between samples, like ‘strain’ or ‘DNA-concentration’.
- Have column names be explanatory, but not have spaces. Use ‘-‘, ‘_’ or camel case instead of a space. For instance ‘library-prep-method’ or ‘LibraryPrep’is better than ‘library preparation method’ or ‘prep’, because computers interpret spaces in particular ways.
- Don’t combine multiple pieces of information in one cell. Sometimes it just seems like one thing, but think if that’s the only way you’ll want to be able to use or sort that data. For example, instead of having a column with species and strain name (e.g. E. coli K12) you would have one column with the species name (E. coli) and another with the strain name (K12). Depending on the type of analysis you want to do, you may even separate the genus and species names into distinct columns.
- Leave the raw data raw - don’t change it!
- Export the cleaned data to a text-based format like CSV (comma-separated values) format. This ensures that anyone can use the data, and is required by most data repositories.
Exercise
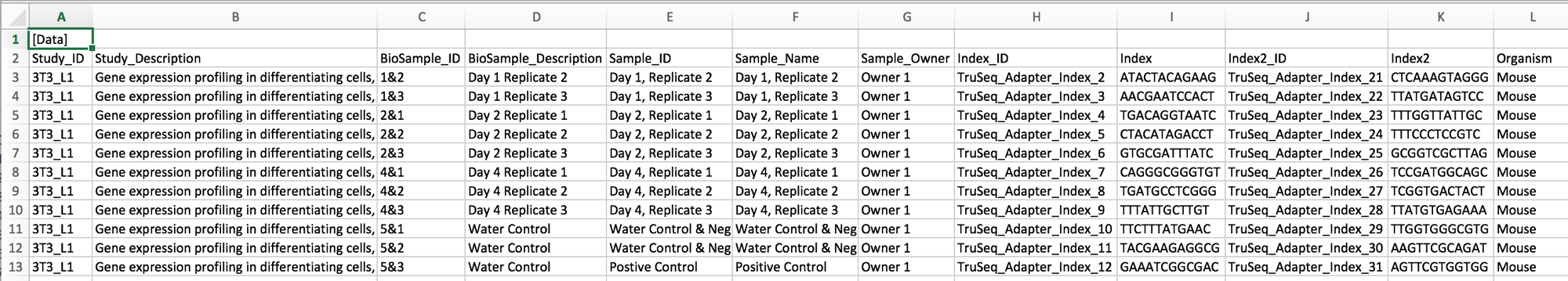
This is some potential spreadsheet data for an experiment being submitted for sequencing. The program bcl2fastq requires this spreadsheet to use as input to demultiplex the sequencing data into separate files, one per sample. With the person next to you, for about 2 minutes, discuss some of the problems with the spreadsheet data shown above.
Solution
A full set of types of issues with spreadsheet data is at the Data Carpentry Ecology spreadsheet lesson. Not all are present in this example. Discuss with the group what they found. The main problem is there are characters in the ids that aren’t allowed, e.g. “,”, “.”, “&” or spaces. Here is a “clean” version of the same spreadsheet:
File and info provided by Dr. Olga Botvinnik at CZ Biohub.
Further notes on data tidiness
Data organization at this point of your experiment will help facilitate your analysis later, as well as prepare your data and notes for data deposition now often required by journals and funding agencies. If this is a collaborative project, as most projects are now, it’s also information that collaborators will need to interpret your data and results and is very useful for communication and efficiency.
Fear not! If you have already started your project, and it’s not set up this way, there are still opportunities to make updates. One of the biggest challenges is tabular data that isn’t formatted so computers can use it, or has inconsistencies that make it hard to analyze.
More practice on how to structure data is outlined in our Data Carpentry Ecology spreadsheet lesson
Tools like OpenRefine can help you clean your data.
Key Points
Metadata is key for you and others to be able to work with your data
Tabular data needs to be structured to be able to work with it effectively